1,200 Genome und 800 Transkriptome
Contact
Project coordinators:
Prof. Tanja Zeller
Prof. Heribert Schunkert
use.access(at)dzhk.de.
Use and Access Office:
Alexandra Klatt
Science Administration/Use and Access Office:
phone: +4930 3465 52910
use.access(at)dzhk.de
About the DZHKomics Resource Project
In this project, the genomes and transcriptomes of subjects from different population-based cohorts were sequenced, generating a dataset that serves as a control resource for further sequencing projects. Thus, the DZHKOmics resource helps to improve our understanding of how genes contribute to the occurrence of disease(s).
Sample preparation, sequencing and processing
In advance of whole genome and RNA sequencing, SOPs for harmonized sample preparation were developed specifically for the DZHKomics resource. This approach was intended to minimize potential confounding influences on the sequencing of samples originating from different cohorts and to achieve high comparability of the generated sequence data.
DNA preparation followed a standardized procedure at the DZHK site in Heidelberg, where whole genome sequencing was also performed on the "HiSeq-X" platform of the 'High Throughput Sequencing Unit'. Data processing was performed in Munich and in Lübeck.
In addition, RNA samples were prepared under standardized conditions in Munich. RNA sequencing was performed on the HiSeq sequencers in Berlin followed by processing in Hamburg.
Subjects
There are 1,300 subjects from six population-based, epidemiological cohort studies in Germany. The sex ratio is almost balanced (48.3% women) and the median age at sample collection is 57 years. Information was collected on various cardiovascular risk factors, which were within the normal range for most subjects. This information is summarized in the DZHKomics phenotype data set, which includes the following variables:
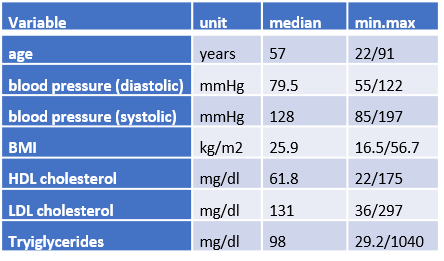
- age
- gender
- height
- weight
- blood pressure (diastolic, systolic)
- HDL and LDL cholesterol
- triglycerides
- smoking status
- diabetes
- family history of heart attack
- familial cases of stroke
- familial cases of CHF/DCM
- medication
Molecular data
Raw DNA sequencing data were processed using established pipelines and software packages (FastQC, bwa-mem, and GATK) according to GATK and gnomAD recommendations. After quality control, genotype data are available for 1099 individuals. A total of approximately 43.9 million variants were identified, including 35.8 million SNVs and 7 million InDels. Information on individual variants including genotype frequencies can be searched and downloaded in our Genome Browser.
Established tools were also used to process the RNA data, including FastQC, STAR, RSEM. Untransformed and normalized count data are available for a total of 775 individuals and 26,914 Ensembl genes.
Cooperation Partner of the DZHKomics Resource
Participating cohorts:
A total of 1,200 whole-genome DNA samples and 800 RNA samples (PaxGene Blood) were sequenced to generate this resource. The sample material was selected from six epidemiological cohort studies already existing at different DZHK member institutions:
- Gutenberg-Gesundheitsstudie (GHS) - Universitätsmedizin der Johannes Gutenberg-Universität Mainz
- Hamburg City Health Study (HCHS) - Universitätsklinikum Hamburg-Eppendorf
- Heidelberg Normal Kontrollen (NOKO) - Universitätsklinikum Heidelberg
- Kooperative Gesundheitsforschung in der Region Augsburg (KORA) - Helmholtz Zentrum München
- Study of Health in Pomerania (SHIP) - Universitätsmedizin Greifswald
- Instituts für Klinische Molekularbiologie Kiel (IKMB) - Universitätsklinikum Schleswig-Holstein

